The recent interest in molecular dynamics (MD) simulations requires for systematic and more precise descriptions of the most commonly used lipids. For example, accurately known lipid areas are crucial when assessing intermolecular interactions taking place within biomembranes. Importantly, the complex structural dynamics found in membranes involve a balance of forces, where area is the product of these forces in the lateral direction (i.e. in-plane). In addition to playing a key role in describing membrane structure and function, knowledge of lateral lipid area is central to molecular dynamics (MD) simulations. We have recently developed a model (i.e. scattering density profile, SDP) to simultaneously analyze x-ray and neutron scattering data from fully hydrated lipid bilayers [Kučerka et al., Biophys. J. 95, 2356 (2008)]. The model is based on volumetric distribution functions that are required to obey spatial conservation, and experimental volume data are incorporated into the analysis (Figure 1). Decisions regarding the specific separation of the sub-molecular components in the model are guided by an MD simulation. Most recently, using this technique we have determined the structural parameters of fluid phase bilayers of common phosphatidylcholines with fully saturated, mixed, and branched fatty acid chains, at several temperatures [Kučerka et al., Biochim. Biophys. Acta 1808, 2761 (2011)]. Bilayer parameters, such as area per lipid and overall bilayer thickness have been obtained in conjunction with intrabilayer structural parameters (e.g. hydrocarbon region thickness).
- Home
-
About
-
Outreach/Education
- Future
-
Science
-
For Users
- Industry
- Publications
-
Instruments
-
High Flux Isotope Reactor
- BIO-SANS | Biological Small-Angle Neutron Scattering Instrument | CG-3
- CTAX | Cold Neutron Triple-Axis Spectrometer | CG-4C
- DEMAND | Dimensional Extreme Magnetic Neutron Diffractometer | HB-3A
- DEV BEAMS | Instrument Development Beamline | HB-2D CG-1A CG-1B CG-4B
- GP-SANS | General-Purpose Small-Angle Neutron Scattering Diffractometer | CG-2
- HIDRA | High Intensity Diffractometer for Residual stress Analysis | HB-2B
- IMAGINE | Laue Diffractometer | CG-4D
- MARS | Multimodal Advanced Radiography Station | CG-1D
- POWDER | Neutron Powder Diffractometer | HB-2A
- PTAX | Polarized Triple-Axis Spectrometer | HB-1
- TAX | Triple-Axis Spectrometer | HB-3
- VERITAS | Versatile Intense Triple-Axis Spectrometer | HB-1A
- WAND² | Wide-Angle Neutron Diffractometer | HB-2C
-
Spallation Neutron Source
- ARCS | Wide Angular-Range Chopper Spectrometer | BL-18
- BASIS | Backscattering Spectrometer | BL-2
- CNCS | Cold Neutron Chopper Spectrometer | BL-5
- CORELLI | Elastic Diffuse Scattering Spectrometer | BL-9
- EQ-SANS | Extended Q-Range Small-Angle Neutron Scattering Diffractometer | BL-6
- FNPB | Fundamental Neutron Physics Beam Line | BL-13
- HYSPEC | Hybrid Spectrometer | BL-14B
- LIQREF | Liquids Reflectometer | BL-4B
- MAGREF | Magnetism Reflectometer | BL-4A
- MANDI | Macromolecular Neutron Diffractometer | BL-11B
- NOMAD | Nanoscale-Ordered Materials Diffractometer | BL-1B
- NSE | Neutron Spin Echo Spectrometer | BL-15
- POWGEN | Powder Diffractometer | BL-11A
- SEQUOIA | Fine-Resolution Fermi Chopper Spectrometer | BL-17
- SNAP | Spallation Neutrons and Pressure Diffractometer | BL-3
- TOPAZ | Single-Crystal Diffractometer | BL-12
- USANS | Ultra-Small-Angle Neutron Scattering Instrument | BL-1A
- VENUS | Versatile Neutron Imaging Instrument | BL-10
- VISION | Vibrational Spectrometer | BL-16B
- VULCAN | Engineering Materials Diffractometer | BL-7
-
High Flux Isotope Reactor
- Staff
Home »
Content »
Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature
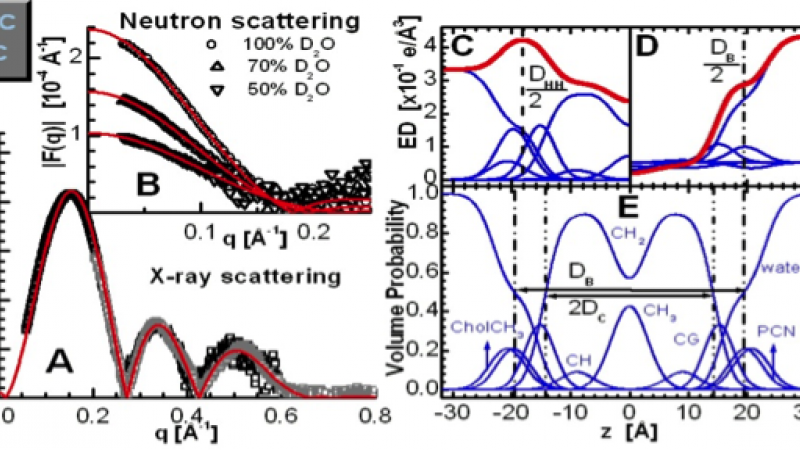
An illustration of lipid bilayer structure determination through the joint refinement of X-ray and neutron scattering data obtained for POPC bilayers at 30oC.
Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature
July 23, 2011
Facility
- Contact Us
Neutron Sciences Directorate
One Bethel Valley Rd
Oak Ridge, TN 37831
Office Phone: 865-574-0558
User Office Phone: 865-574-4600
One Bethel Valley Rd
Oak Ridge, TN 37831
Office Phone: 865-574-0558
User Office Phone: 865-574-4600
QUICK LINKS
Oak Ridge National Laboratory is managed by UT-Battelle LLC for the US Department of Energy