Molecular Structure of Sphingomyelin (SM) Bilayers
Scientific Achievement
The molecular structure of SM bilayers was determined, providing a quantitative measure of the area per lipid.
Significance and Impact
The results enable an accurate refinement of the force fields used in simulations that are crucial for the study of membrane systems in general.
Research Details
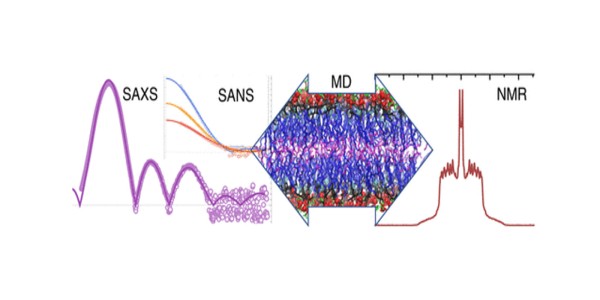
- The structures of SM bilayers were measured by small angle scattering of X-rays (SAXS) and neutrons (SANS), with SANS providing the crucial multiple contrast data sets needed to refine the relevant parameters.
- The data was analyzed using a novel scattering density profile method.
- The SMs were also characterized by differential scanning calorimetry, nuclear magnetic resonance (NMR), and electron spin resonance.
- Bilayers were further analyzed with unconstrained and area-constrained molecular dynamics (MD) simulations.
“Molecular Structure of Sphingomyelin in Fluid Phase Bilayers Determined by the Joint Analysis of Small-Angle Neutron and X‑ray Scattering Data,”
Milka Doktorova, Norbert Kučerka, Jacob J. Kinnun, Jianjun Pan, Drew Marquardt, Haden L. Scott, Richard M. Venable, Richard W. Pastor, Stephen R. Wassall, John Katsaras, and Frederick A. Heberle,
Journal of Physical Chemistry B 124, 5186-5200 (2020).
DOI: https://doi.org/10.1021/acs.jpcb.0c03389